Thank you professor for the response!
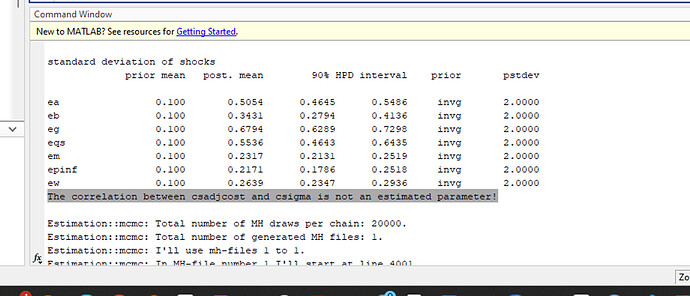
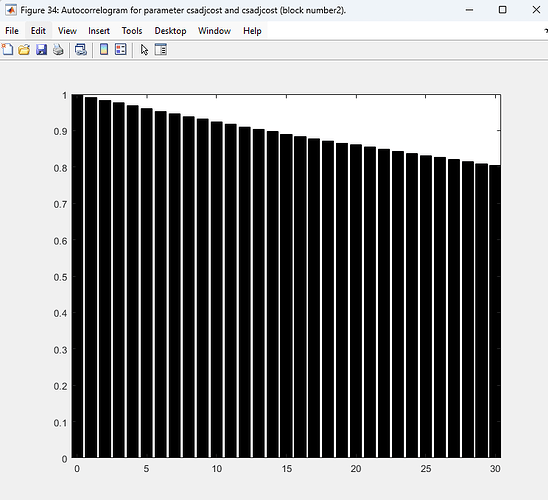
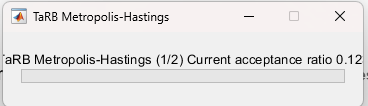
Re 1(and 2) The autocorrelogram code works now, but the problem is that most parameters have over 0.8-0.9 autocorrelation, way too high, aligning with the problematic acceptance rate (>0.5).
As you suggested in 2., I thought maybe the number of samples were insufficient, so I increased the number of simulations per block from 20,000 to 60,000( i have 2 blocks), but there was barely no change in the acceptance ratio and autocorrelogram… Do you think it is ok to conclude from this that SW 2007 is ill-posed for Bayesian computation? Are there any other approaches I might take?
Re 3[First point].
Thank you, I am wondering then if the degeneracy might have something to do with the way I’m setting up the parameters in dynare?
For example, I get the below warning every time I run the code that your group has provided for SW 2007:
Starting Dynare (version 5.5).
Calling Dynare with arguments: none
Starting preprocessing of the model file ...
WARNING: in the 'steady_state_model' block, variable 'ewma' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'epinfma' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'zcapf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'rkf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'kf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'pkf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'cf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'invef' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'yf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'labf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'wf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'rrf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'mc' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'zcap' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'rk' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'k' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'pk' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'c' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'inve' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'y' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'lab' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'pinf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'w' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'r' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'a' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'b' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'g' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'qs' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'ms' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'spinf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'sw' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'kpf' is not assigned a value
WARNING: in the 'steady_state_model' block, variable 'kp' is not assigned a value
Found 40 equation(s).
Evaluating expressions...done
Computing static model derivatives (order 1).
Computing dynamic model derivatives (order 2).
Processing outputs ...
done
Preprocessing completed.
You did not declare endogenous variables after the estimation/calib_smoother command.
[�Warning: Some of the parameters have no value (ccs, cinvs, crdpi) when using initial_estimation_checks. If these
parameters are not initialized in a steadystate file or a steady_state_model-block, Dynare may not be able to solve the
model. Note that simul, perfect_foresight_setup, and perfect_foresight_solver do not automatically call the steady state
file.]�
Initial value of the log posterior (or likelihood): -1484.2789
Re 3[2nd point].Relatedly, to dive into this issue in a more tractable way, I am starting with your suggested RBC_baseline material DSGE_mod/RBC_baseline at master · JohannesPfeifer/DSGE_mod · GitHub
and trying to get multiple draws from the posterior as well as the likelihood evaluated at that parameter value by using the dsge_likelihood function, but the problem I have now is I dont have a way to see the “info” variable from the dsge_likelihood (which is important because that output shows whether the kalman filter’s matrices are nonsingular or not).
Specifically,
as if I use the
posterior_function(function=‘OOI_and_likelihood’,sampling_draws=100) as a wrapper, I can’t seem to be able to recover the “info” output,
and on the other hand, if I
add the code in the attached file “RBC_baseline_first_diff_bayesian.mod”(I added some lines to your original code),
[fval, info] = dsge_likelihood(xparam1,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,prior_bounds(bayestopt_, options_.prior_trunc),oo_)
returns the error
Unrecognized function or variable 'xparam1'.
Error in RBC_baseline_first_diff_bayesian.driver (line 329)
[fval, info] = dsge_likelihood(xparam1,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,prior_bounds(bayestopt_, options_.prior_trunc),oo_)
Error in dynare (line 281)
evalin('base',[fname '.driver']);
Error in yp_script_basic (line 3)
dynare RBC_baseline_first_diff_bayesian.mod
although in examples I saw online, the xparam1 was being used.
Thank you so much for your time!