Hi,
I’m new at Dynare trying to replicate a code of a paper. I want to add Bayesian estimation but something went wrong.
dynare sw_2020.mod
Starting Dynare (version 5.1).
Calling Dynare with arguments: none
Starting preprocessing of the model file ...
Found 61 equation(s).
Evaluating expressions...done
Computing static model derivatives (order 1).
Computing dynamic model derivatives (order 1).
Processing outputs ...
done
Preprocessing completed.
STEADY-STATE RESULTS:
Y 2.89875
C 1.71006
I 0.608939
G 0.579751
L 1
Ld 1
Ih 0.608939
Yw 2.89875
K 24.3576
u 1
w 1.75682
wr 1.75682
mrs 1.59711
Pi 1
Pir 1
pk 1
pw 0.909091
f1 3096.91
f2 1939.07
x1 10.3851
x2 11.4237
vp 1
vw 1
Q 26.6813
QB 30.7424
RF 1.01248
RB 1.00753
Rd 1.00503
Rre 1.00503
Rtr 1.00503
M1 1.24827
M2 1.2011
mu 0.596469
Lam 0.995
fw 0.739456
f 0.738369
b 0.0386595
re 0.289875
d 16.9432
n 4.2358
lambda 0.0351955
phi 4.7452
Omega 3.01855
fcb 0.00108644
bcb 0.00848624
bG 0.0471458
A 1
theta 0.658515
logY 1.06428
logC 0.536531
logI -0.496037
logLd 0
logPi 0
logRd 2.00502
exr 2.95588
logQ 3.28396
C_obs 0
Y_obs 0
I_obs 0
Pi_obs 0
Rd_obs -1.45347
You did not declare endogenous variables after the estimation/calib_smoother command.
Posterior IRFs, smoothed variables will be computed for the 61
endogenous variables of your model, this can take a long
time ....
Choose one of the following options:
[1] Consider all the endogenous variables.
[2] Consider all the observed endogenous variables.
[3] Stop Dynare and change the mod file.
[4] Consider all the endogenous and auxiliary variables.
options [default is 1] = 1
Initial value of the log posterior (or likelihood): -77566642549.1891
==========================================================
Change in the posterior covariance matrix = 1.
Change in the posterior mean = 0.96.
Current mode = 33978768.7551
Mode improvement = 77532663780.434
New value of jscale = 5.8706e-08
==========================================================
==========================================================
Change in the posterior covariance matrix = 5.378e-11.
Change in the posterior mean = 0.90175.
Current mode = 37713413.4273
Mode improvement = 3734644.6723
New value of jscale = 4.2326e-08
==========================================================
错误使用 chol
矩阵必须为正定矩阵。
出错 gmhmaxlik_core (第 194 行)
dd = transpose(chol(CovJump));
出错 gmhmaxlik (第 100 行)
[PostMode, PostVariance, Scale, PostMean] = gmhmaxlik_core(fun, OldPostMode, bounds, gmhmaxlikOptions, Scale, flag, MeanPar, OldPostVariance, varargin{:});
出错 dynare_minimize_objective (第 336 行)
[opt_par_values, hessian_mat, Scale, fval] = gmhmaxlik(objective_function, start_par_value, ...
出错 dynare_estimation_1 (第 211 行)
[xparam1, fval, exitflag, hh, options_, Scale, new_rat_hess_info] = dynare_minimize_objective(objective_function,xparam1,options_.mode_compute,options_,[bounds.lb bounds.ub],bayestopt_.name,bayestopt_,hh,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,bounds,oo_);
出错 dynare_estimation (第 118 行)
dynare_estimation_1(var_list,dname);
出错 sw_2020.driver (第 894 行)
oo_recursive_=dynare_estimation(var_list_);
出错 dynare (第 281 行)
evalin('base',[fname '.driver']);
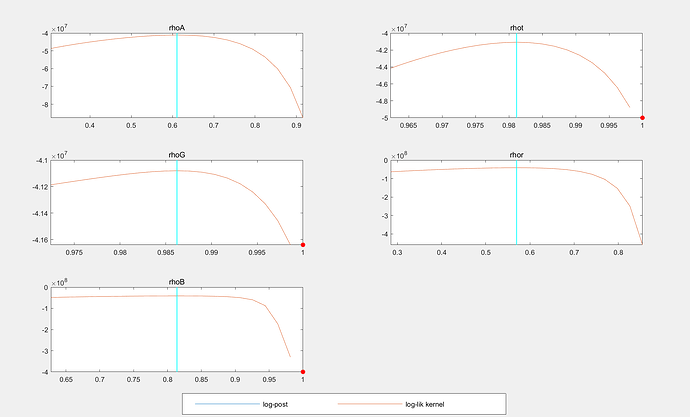
If the estimated_params only have
rhoA, 0.9 , 0 ,2, beta_pdf, 0.80 , 0.10;
rhot, 0.9, 0 ,2, beta_pdf, 0.80 , 0.10;
rhoG, 0.9 , 0 ,2, beta_pdf, 0.80 , 0.10;
rhor, 0.9 , 0 ,2, beta_pdf, 0.80 , 0.10;
rhoB, 0.9, 0 ,2, beta_pdf, 0.80 , 0.10;
the code can be run. Then the mode_check_plots looks like:
I don’t know how to tackle the problem, please help me.
sw_2020.mod (9.4 KB)
data_123.m (3.0 KB)
+sw_2020.rar (23.3 KB)