Thanks again @jpfeifer , I do appreciate your time. So here are my updated questions / concerns after running my code and comparing it with the Atkinson et al (2020 JME) example.
1) In both my model and the Atkinson et al. example, I set mode_compute=4 and mh_replic=400000. I try to obtain the _mode.mat file from the Atkinson et al. example without using their saved one. Running either of these models with mh_replic>0 produces on the one hand some reasonable posterior values as well as mode_check figures but fails to produce a new _mode.mat file on the other. The error message received in both models, after the posteriors and figures are produced, is:
**""**Log data density [Laplace approximation] is NaN.
Error using chol
Matrix must be positive definite.
Error in posterior_sampler_initialization (line 77)
d = chol(vv);
Error in posterior_sampler (line 60)
posterior_sampler_initialization(TargetFun, xparam1, vv, mh_bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_, dispString);
Error in dynare_estimation_1 (line 432)
posterior_sampler(objective_function,posterior_sampler_options.proposal_distribution,xparam1,posterior_sampler_options,bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_,dispString);
Error in dynare_estimation (line 105)
dynare_estimation_1(var_list,dname);
Error in NKM.driver (line 947)
oo_recursive_=dynare_estimation(var_list_);
Error in dynare (line 310)
evalin('base',[fname '.driver']);**""**
Setting mh_replic=0 and mode_compute=4 does not produce this error message but still produces NaN for the Laplace. Given that this is a shared problem with both the official example and my code, I’m not sure what could be wrong here.
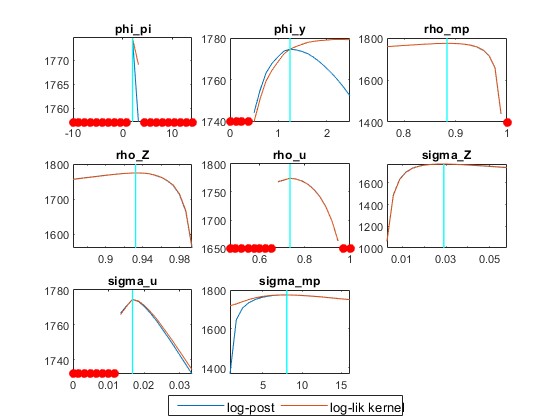
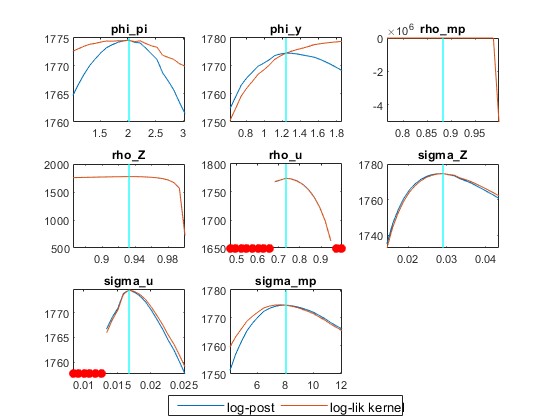
2) How important is using a 0 value for the prior_trunc? I understand that it deals with unit roots but having prior_trunc=0 or ignoring this option in the estimation command produces slightly different results, especially the mode_check figure for the inflation response in the Taylor rule that is truncated very close to its prior. Should this option be used or not in general? For example, In the Atkinson et al. example, with mode_compute=4 and mh_replic=400000, the model fails to produce the full mode check plots when ignoring prior_trunc=0.
- Beyond the code for my .mod model file and Data and Data 2 .mat files, which are interlinked, I attach a picture of the mode_check figure produced from my model for both prior_trunc=0 and no prior_trunc (I’m not sure why you referred to the ‘DataNL.mat’ file in the previous message?). In any case, I’m not sure how to interpret the mode_check files, especially thi phi_pi with prior_trunc=0 and the rho_mp without the prior_trunc option.
Apologies for the long message, I tried to be as elaborate and clear as possible. I hope you can point me in the right direction for my three concerns above. Thanks in advance and all the best.
NKModelNL.mod (6.6 KB)
Data.mat (3.3 KB)
Data2.mat (3.2 KB)