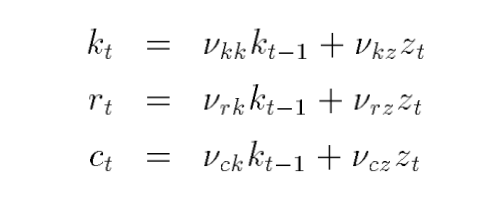Hi all,
I’m exploring Dynare’s filtering capabilities through a simple exercise: I take a basic state-space model, apply MATLAB’s smoother to recover the smoothed states, and compare the results to those produced by Dynare. The state-space model is based on the policy function of a neoclassical growth model, using the calibration provided on page 19 of Uhlig’s toolkit, and assumes no measurement error and has one 1 observable (PCE). The data file for Dynare is log_deviation_data.csv.
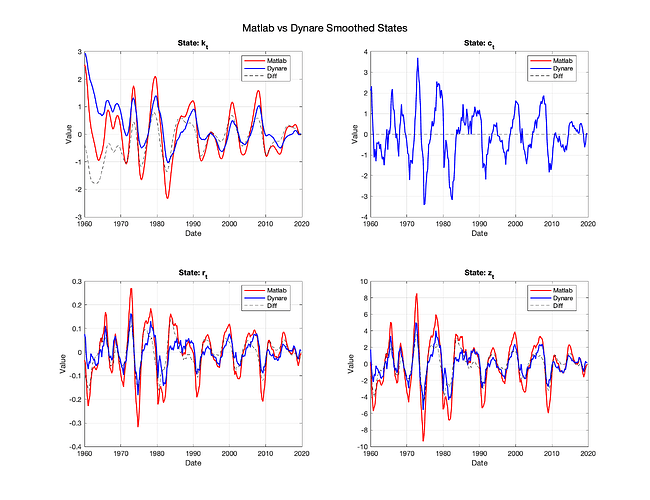
I provide Dynare with the same model and calibration, using “mode_compute=0” to bypass estimation. I also give the two models the same initial assumptions for the states (S_0 and P_0). However, aside from the one observed state, I get noticeable differences between the series produced by MATLAB and those generated by Dynare (see image below).
The files “example_ssm.m and ParamMap.m” generate the smoothed series using Matlab’s KS. The file “try_1.mod” is Dynare’s mod file. Finally, “try1_comp.m” compares results and plots the series and their difference. I’m running Dynare 5.5:
log_deviation_data.csv (4.3 KB)
example_ssm2.m (4.0 KB)
ParamMap.m (1.0 KB)
try1_comp.m (1.3 KB)
try_1.mod (2.0 KB)
The problem appears to be the transition matrix A, where it should be:
A = [ 0.965, 0, 0, 0.07125;
0.618, 0, 0, 0.28975;
-0.022, 0, 0, 0.03325;
0, 0, 0, 0.950];
(as in the Matlab file), in Dynare it is being populated as:
A = [ 0.965, 0, 0, 0.07125;
0, 0.618, 0, 0.28975;
0, 0, -0.022, 0.03325;
0, 0, 0, 0.950];
I guess that I’m overlooking something in the model description, but I haven’t been able to identify what it is. Any help would be greatly appreciated @jpfeifer. Thank you!