Thanks a lot for the reply.
-
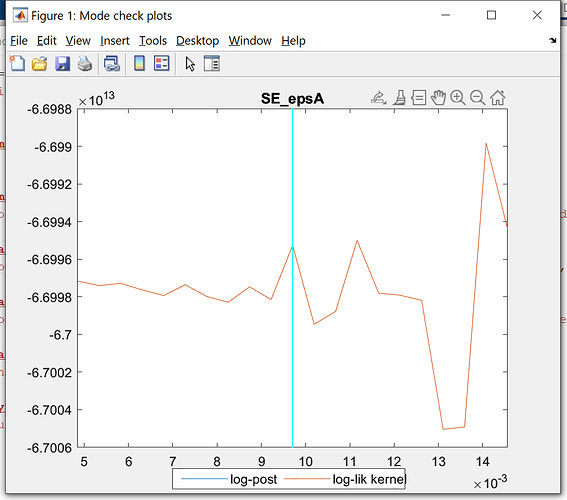
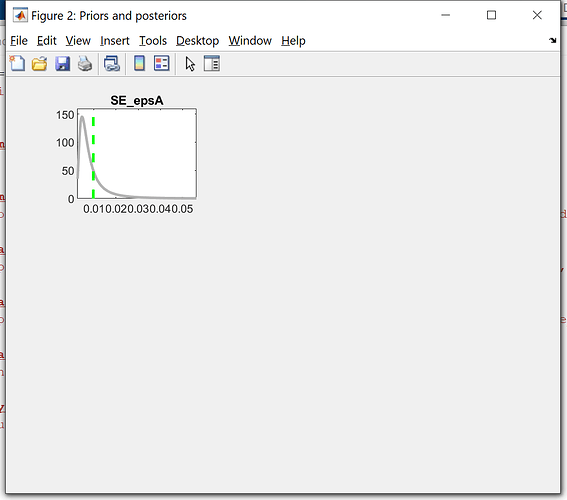
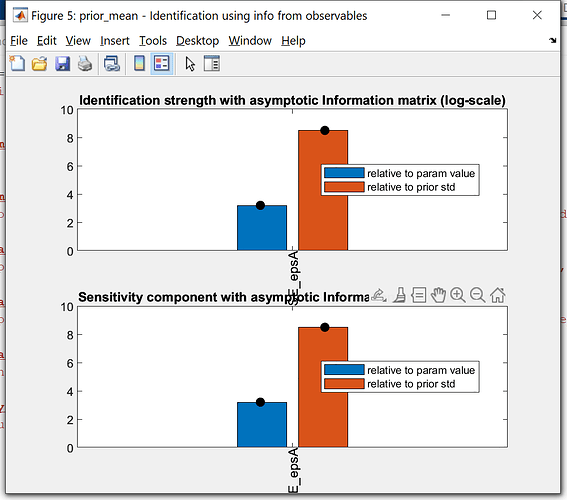
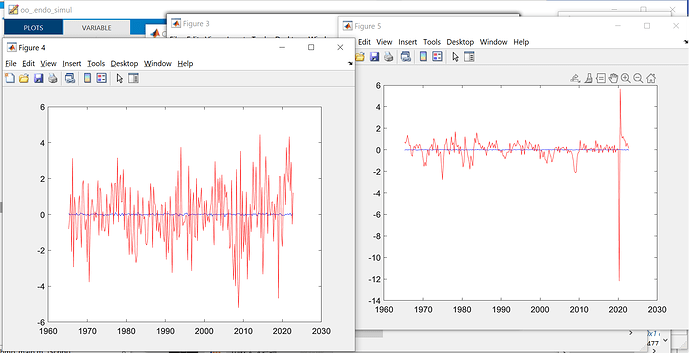
Just want to double check with you, that the simulation data of the model is the separate different first-period irf of iid shocks combined together.
If this is the case, I am just curious, why compare the detrended empirical data with the irf of the first period, why not the first 5 or 10, or the second. Is there any autocorrelation considered in the simulation data from model. May I know if there is any theory backing this simulation or comparison.
-
I have some problem using fsolve in the steady_state_model block.
I read this in the dynare manual:
It says I can assign several variables at the same time, but I am not sure how to write it, have problem doing this.
My code is the following:
steady_state_model;
…
[Qh,Nc] = ss2var(alfa,delttass,Ass,rss,gss,hb,deltah,ksi,epsilonass,Qh0,Nc0);
…
end;
ss2var is the steady state calculation function as the following:
function XX_ss=ss2var(alfa,delttass,Ass,rss,gss,hb,deltah,ksi,epsilonass,Qh0, Nc0)
options=optimset(‘Display’,‘Final’,‘TolX’,1e-10,‘TolFun’,1e-10);
[XX_ss,fval,exitf] = fsolve(@(XX)f_qhncsp(XX,alfa,delttass,Ass,rss,gss,hb,deltah,ksi,epsilonass),[Qh0 Nc0]);
f_qhncsp will return the value of Qh and Nc as:
function diff=f_qhncsp(XX,alfa,delttass,Ass,rss,gss,hb,deltah,ksi,epsilonass)
Qh = XX(1);
Nc = XX(2);
…
diff(1) = delttass/(1-N)-W/Ct;
diff(2) = QhH - gamCt/(1-(1-deltah)/R);
it returns:
Processing outputs …
done
Preprocessing completed.
Error using ss2var
Too many output arguments.
Error in hm72fsdraft.steadystate (line 15)
[ys_(8),ys_(13)]=ss2var(params(1),params(2),params(3),params(4),params(5),params(6),params(7),params(8),params(9),params(43),params(44));
Error in evaluate_steady_state_file (line 53)
[ys,params1,check] = h_steadystate(ys_init, exo_ss, params);
Error in evaluate_steady_state (line 254)
[ys,params,info] = evaluate_steady_state_file(ys_init,exo_ss,M, options,steadystate_check_flag);
Error in resid (line 66)
evaluate_steady_state(oo_.steady_state,M_,options_,oo_,0);
Error in hm72fsdraft.driver (line 789)
resid;
Error in dynare (line 281)
evalin(‘base’,[fname ‘.driver’]);
Thanks a lot.
hm72fsdraft.mod (8.3 KB)
ss2var.m (263 Bytes)
f_qhncsp.m (1.3 KB)