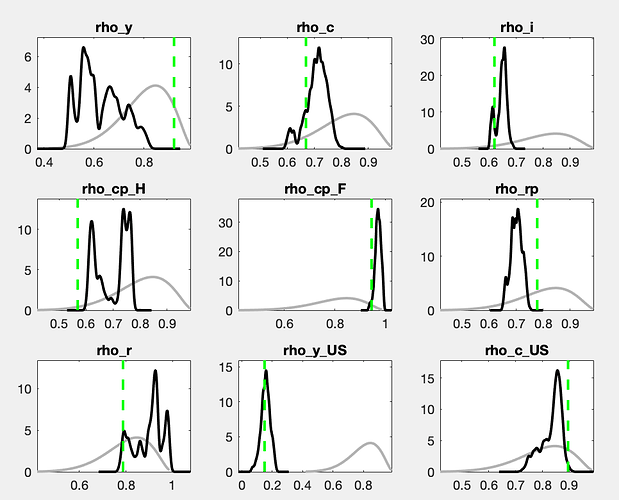
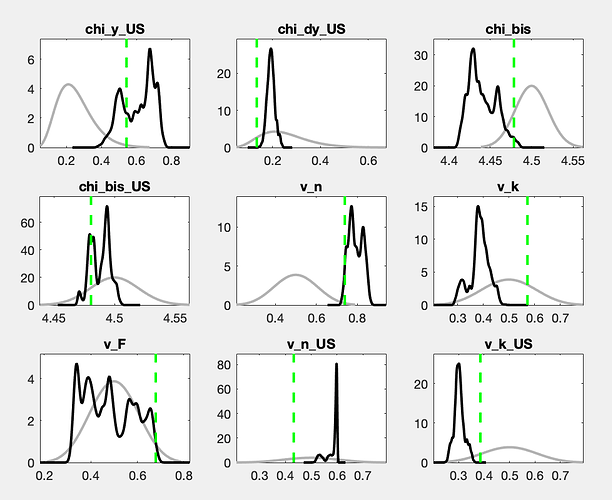
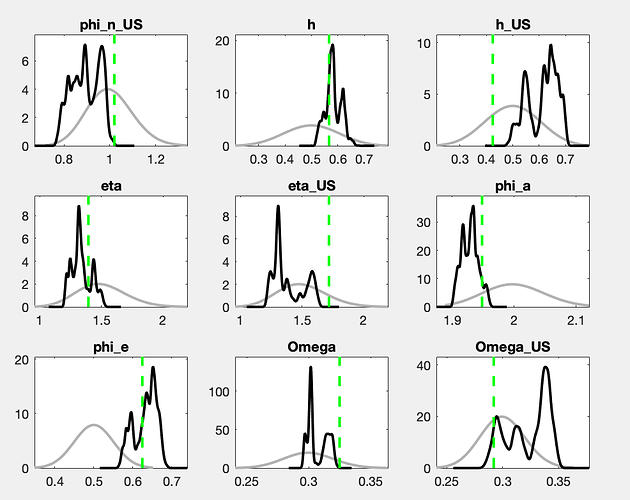
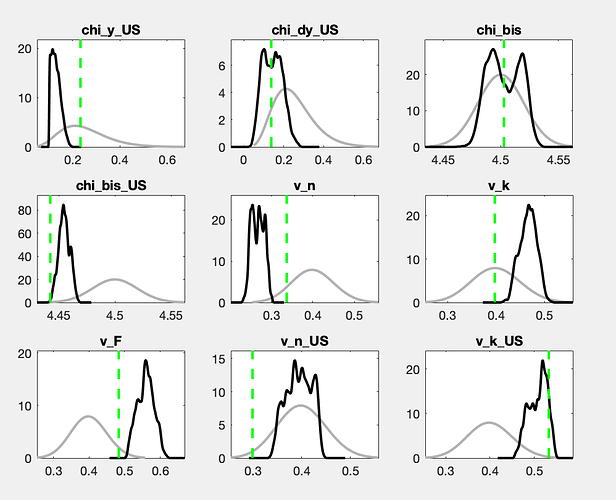
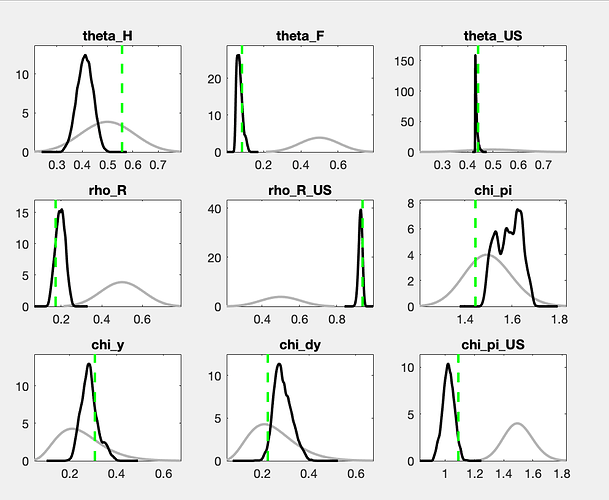
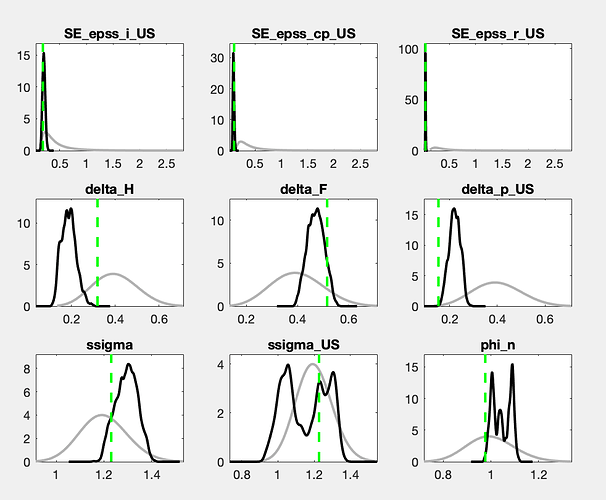
There is still a considerable drift in the parameters. How does the posterior evolve? And what are the parameters drifting here?
The posteriors looks worse than before, could that be related to the burn in ? I’ll re-run it next week.
With a drift like the one shown in the trace plots, a burn-in will not matter much.
What are your thoughts of posteriors like these?
I would say the worst are ssigma_US and phi_n, the rest looks better (some have a mode and posterior that differs, is this important?)
I’m estimating 57 parameters in my model, I increased the number of draws to 3 million which took 21h to compute on my computer. I don’t really think I can increase it anymore, and I don’t know if any examiner is going to accept codes that takes that long to run?
Also, the acceptance rate of my chains are only 25% percent, I’ll try to increase that, but can this significantly change the posteriors?
What would be your verdict if you were my supervisor/examiner? Is this acceptable results to claim convergece of the estimation in a master thesis?
Thanks a lot for all your time, I’ll acknowledge that in my thesis.
The chains still do not look like convergence. With a lot of fantasy, you can imagine additional draws to fill out the valleys of the shown histograms. If this were for a paper to be published, I would advocate for either a lot more draws or the use of a different sampler (like TaRB-MH or slice).
For a thesis, you have to ask your adviser. It may be beyond his expectations to fix this.
I get the following error when using TaRB
"Error using chol_SE (line 74)
A is not symmetric
Error in posterior_sampler_iteration (line 114)
proposal_covariance_Cholesky_decomposition_upper=chol_SE(inverse_hessian_mat,0);
Error in posterior_sampler_core (line 197)
[par, logpost, accepted, neval] = posterior_sampler_iteration(TargetFun, last_draw(curr_block,:), last_posterior(curr_block), sampler_options,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,mh_bounds,oo_);
Error in posterior_sampler (line 121)
fout = posterior_sampler_core(localVars, fblck, nblck, 0);
Error in dynare_estimation_1 (line 471)
posterior_sampler(objective_function,posterior_sampler_options.proposal_distribution,xparam1,posterior_sampler_options,bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_);
Error in dynare_estimation (line 118)
dynare_estimation_1(var_list,dname);
Error in model.driver (line 807)
oo_recursive_=dynare_estimation(var_list_);
Error in dynare (line 281)
evalin(‘base’,[fname ‘.driver’]);"
Haven’t tried the slice sampler yet, but I can see from the manual that it’s incompatible with prior_trunc=0.
Could you please provide me with the file to replicate the issue?
It happens after the mode is found, in the beginning of the first chain.
dataset1.mat (8.7 KB)
model.mod (8.2 KB)
I am still experimenting with your codes as I could not replicate the crash yet. That being said, I noticed that mode_compute=6 failed to find the true mode. That may explain the drift in the chains.
Also, slice needs bounded prior. Therefore, prior_trunc=0 does not work, but you could simply set the truncation to a small number.
Is the TaRB working without any changes to my code? Why would I get an error approx 1h into the runtime? I’m using Dynare 5.0.
I am still experimenting with your codes as I could not replicate the crash yet. That being said, I noticed that
mode_compute=6failed to find the true mode. That may explain the drift in the chains.
Hmm, it didn’t really crash, but the sampling froze in the beginning of the first chain and I got the error message stated above, after some hour I simply quit Matlab. Why would mode_compute=6 fail? I did try default 4 and 9 but it didn’t work so I simply choose 6 since it’s last resort.
Also, slice needs bounded prior. Therefore,
prior_trunc=0does not work, but you could simply set the truncation to a small number.
I might look into that, right now I’m testing 10m draws with the random walk MH. But if mode_compue=6 doesn’t work properly, I will still get faulty posterior distributions?
Sincerely,
I loaded the mode-file from mode_compute=6 and used mode_compute=5. It found a region of considerably higher posterior density. The issue I encounter then is the various prior bounds you imposed.
I’ll try and remove the prior bounds then, they are not important actually. But will mode_compute=5 work without loading a mode-file? If mode_compute=6 fails to find the true mode, then I need another optimizer for the mode computation, correct?
This is the mode I found after relaxing some of the bounds. mode_compute=5 often works well.
model_TaRB_mode.mat (46.4 KB)
I got this error when removing all the bounds and mode_compute=5.
"Log data density [Laplace approximation] is NaN.
Error using chol
Matrix must be positive definite.
Error in posterior_sampler_initialization (line 84)
d = chol(vv);
Error in posterior_sampler (line 60)
posterior_sampler_initialization(TargetFun, xparam1, vv, mh_bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_);
Error in dynare_estimation_1 (line 471)
posterior_sampler(objective_function,posterior_sampler_options.proposal_distribution,xparam1,posterior_sampler_options,bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_);
Error in dynare_estimation (line 118)
dynare_estimation_1(var_list,dname);
Error in model.driver (line 806)
oo_recursive_=dynare_estimation(var_list_);
Error in dynare (line 281)
evalin(‘base’,[fname ‘.driver’]);"
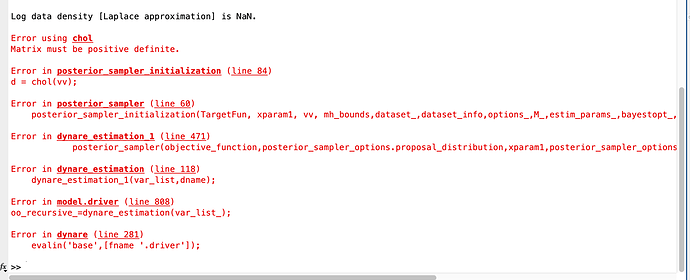
I also tried the mode file you provided, didn’t work, same error see picture.
What could possible be the reason for this?
You need to provide the full error message, not just the last bit. Usually, there is some relevant information before it. You can also upload the log-file.
Initial value of the log posterior (or likelihood): 1407.1851
POSTERIOR KERNEL OPTIMIZATION PROBLEM!
(minus) the hessian matrix at the "mode" is not positive definite!
=> posterior variance of the estimated parameters are not positive.
You should try to change the initial values of the parameters using
the estimated_params_init block, or use another optimization routine.
Warning: The results below are most likely wrong!
> In dynare_estimation_1 (line 308)
In dynare_estimation (line 118)
In model.driver (line 808)
In dynare (line 281)
MODE CHECK
Fval obtained by the minimization routine (minus the posterior/likelihood)): -1407.185109
Warning: Matrix is singular, close to singular or badly scaled. Results may be inaccurate. RCOND = NaN.
> In dynare_estimation_1 (line 331)
In dynare_estimation (line 118)
In model.driver (line 808)
In dynare (line 281)
RESULTS FROM POSTERIOR ESTIMATION
parameters
prior mean mode s.d. prior pstdev
delta_H 0.4000 0.0943 NaN beta 0.1000
delta_F 0.4000 0.3025 NaN beta 0.1000
delta_p_US 0.4000 0.1376 NaN beta 0.1000
ssigma 1.2000 0.7393 NaN gamm 0.1000
ssigma_US 1.2000 1.2885 NaN gamm 0.1000
phi_n 1.0000 0.8182 NaN gamm 0.1000
phi_n_US 1.0000 1.0436 NaN gamm 0.1000
h 0.5000 0.1839 NaN beta 0.1000
h_US 0.5000 0.9840 NaN beta 0.1000
eta 1.5000 0.9598 NaN gamm 0.2000
eta_US 1.5000 0.3708 NaN gamm 0.2000
phi_a 2.0000 1.9960 NaN invg 0.0500
phi_e 0.5000 0.4408 NaN beta 0.0500
Omega 0.3000 0.3076 NaN beta 0.0200
Omega_US 0.3000 0.3238 NaN beta 0.0200
theta_H 0.5000 0.1486 NaN beta 0.1000
theta_F 0.5000 0.0678 NaN beta 0.1000
theta_US 0.5000 0.4300 NaN beta 0.1000
rho_R 0.5000 0.2412 NaN beta 0.1000
rho_R_US 0.5000 0.9593 NaN beta 0.1000
chi_pi 1.5000 1.7040 NaN gamm 0.1000
chi_y 0.2500 1.3922 NaN gamm 0.1000
chi_dy 0.2500 0.4075 NaN gamm 0.1000
chi_pi_US 1.5000 1.3833 NaN gamm 0.1000
chi_y_US 0.2500 0.3376 NaN gamm 0.1000
chi_dy_US 0.2500 0.2210 NaN gamm 0.1000
chi_bis 4.5000 4.4993 NaN gamm 0.0200
chi_bis_US 4.5000 4.5044 NaN gamm 0.0200
v_n 0.4000 0.3835 NaN beta 0.0500
v_k 0.4000 0.3959 NaN beta 0.0500
v_F 0.4000 0.3839 NaN beta 0.0500
v_n_US 0.4000 0.4294 NaN beta 0.0500
v_k_US 0.4000 0.4431 NaN beta 0.0500
rho_y 0.8000 0.9737 NaN beta 0.1000
rho_c 0.8000 0.9930 NaN beta 0.1000
rho_i 0.8000 0.1370 NaN beta 0.1000
rho_cp_H 0.8000 0.9041 NaN beta 0.1000
rho_cp_F 0.8000 0.2270 NaN beta 0.1000
rho_rp 0.8000 0.9431 NaN beta 0.1000
rho_r 0.8000 0.9493 NaN beta 0.1000
rho_y_US 0.8000 0.2803 NaN beta 0.1000
rho_c_US 0.8000 0.2461 NaN beta 0.1000
rho_i_US 0.8000 0.7406 NaN beta 0.1000
rho_cp_US 0.8000 0.4912 NaN beta 0.1000
rho_r_US 0.8000 0.6051 NaN beta 0.1000
standard deviation of shocks
prior mean mode s.d. prior pstdev
epss_y 0.5000 0.0586 NaN invg Inf
epss_c 0.5000 0.1694 NaN invg Inf
epss_i 0.5000 0.2392 NaN invg Inf
epss_cp_H 0.5000 0.1502 NaN invg Inf
epss_cp_F 0.5000 1.2066 NaN invg Inf
epss_rp 0.5000 0.1945 NaN invg Inf
epss_r 0.5000 0.0499 NaN invg Inf
epss_y_US 0.5000 0.2477 NaN invg Inf
epss_c_US 0.5000 1.7716 NaN invg Inf
epss_i_US 0.5000 0.2355 NaN invg Inf
epss_cp_US 0.5000 0.1421 NaN invg Inf
epss_r_US 0.5000 0.0443 NaN invg Inf
Log data density [Laplace approximation] is NaN.
Error using chol
Matrix must be positive definite.
Error in posterior_sampler_initialization (line 84)
d = chol(vv);
Error in posterior_sampler (line 60)
posterior_sampler_initialization(TargetFun, xparam1, vv, mh_bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_);
Error in dynare_estimation_1 (line 471)
posterior_sampler(objective_function,posterior_sampler_options.proposal_distribution,xparam1,posterior_sampler_options,bounds,dataset_,dataset_info,options_,M_,estim_params_,bayestopt_,oo_);
Error in dynare_estimation (line 118)
dynare_estimation_1(var_list,dname);
Error in model.driver (line 808)
oo_recursive_=dynare_estimation(var_list_);
Error in dynare (line 281)
evalin('base',[fname '.driver']);
I’m using the mode file you provided me.
You can verify that theta_US is at its bound. You told me you wanted to remove them.
You can verify that
theta_USis at its bound. You told me you wanted to remove them.
How do I verify theta_US? Just get this message: “Error using check_prior_bounds (line 39)
Initial value(s) of theta_US are outside parameter bounds. Potentially, you should set prior_trunc=0. If you used the
mode_file-option, check whether your mode-file is consistent with the priors.”
The bounds doesn’t matter for me if the model works, but it didn’t. I tried remove all bounds, change mode_compute=5 and using the TaRB. Error messages as just stated. Then tried the mode file you provided and same errors. Now I’m trying to put back some since you said you relaxed some of the bounds but not all? And it worked?
Try these two. After mode_compute=5, I ran mode_compute=6:
model_TaRB_mode.mat (46.7 KB)
model_TaRB.mod (8.1 KB)
They seem to work, thank you.
A couple of questions:
-
Why didn’t mode_compute=6 find the true mode initially? Is this common?
-
Why did mode_compute=5 and then =6 work?
-
I’ll run two more models, the second is the same but where I model the shock processes differently, meaning some priors/parameters will have to be changed/ added. The other model will also be the same as the first one, but without any frictions. I guess the mode file just provided won’t work for these two models, how can I solve this issue? Since mode_compute=5 won’t work (I’ll try first with the mod file you just updated).
-
Regarding the TaRB, I assume the principle regarding number of chains is the same across samplers? Like two chains is standard and fine? No reason to change? I still need to adjust the jscale to get around 1/3 acceptance ratio as usually? Also, I’ve seen researchers use around 10 000 draws with the TaRB, I guess that will be fine in this case as well?
Thank you for all your time. Highly appreciated.